The machine assisted product screening (MAPS) project is dedicated to leveraging the collective biological and chemical knowledge to bridge the gap between synthetic biology and synthetic chemistry. Its primary aim is to explore bio-derived monomers for creating sustainable polymers with customizable properties. With the pressing need to combat pollution caused by petroleum-based plastics, MAPS will provide a vital platform for researchers to discover and develop promising alternatives.
A few pre-existing machine learning (ML) tools have excelled in predicting chemical reactions based on their reactivities. However, when it comes to polymerization reactions involving radicals and other complex processes, these conventional tools often fall short, predicting unrealistic outcomes. This limitation arises from the lack of training data specific to polymer synthesis.
To address this challenge, MAPS is on a mission to compile a comprehensive database of polymerizability tailored to natural product molecules. Establishing the MAPS database is a big challenge due to the absence of a pre-existing database in this domain. To address this obstacle, we are actively seeking synthetic expertise from experts to develop a targeted ML algorithm for predicting polymerizability. But we can't do it alone...
By actively participating in the MAPS project, you're not just contributing data – you're driving sustainable materials innovation in polymer research. Join us in shaping a sustainable future and innovating bio-friendly materials for polymer community!
Are you ready to join us in enriching our MAPS database?
A step-by-step guide to contributing to sustainable polymer innovation:
First, log into the BioPACIFIC MIP portal and visit the MAPS 2.0 web application (step 1). Create an account here.
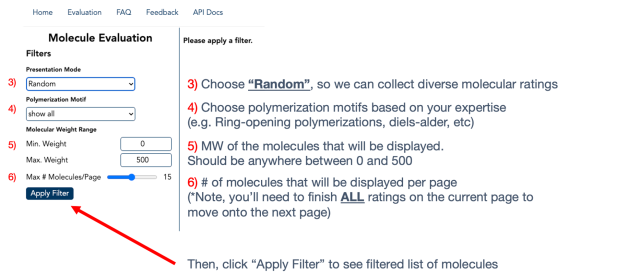
Next, navigate to the "Evaluation" tab (step 2).

Apply filters (steps 3-6) to rate molecules and functional groups based on each polymerization approach. Molecules shown will be sorted by their substructures.
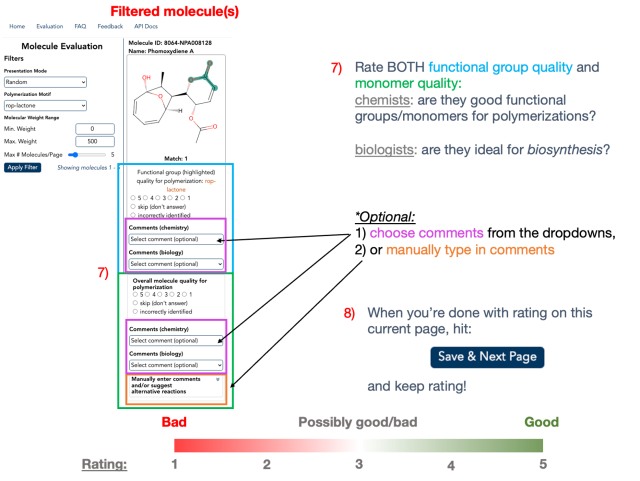
Now is the fun part! Assume that you’re rating the molecular/monomer quality and functional group quality as if you’re rating a restaurant in Yelp (step 7)! The rating scale spans from 1 (“bad”) through 5 (“good”). If the molecule shown is not ideal as-is, but has the potential to become a promising molecule with slight modifications, you can rate them as 3 (“intermediate”). Relevant/helpful comments can be chosen from the dropdowns. If you can’t find an option you like, you can also manually type in your comments.
After completing the ratings on the current page, click "Save & Next Page" to record your responses (step 8).
Contact
Keep up the momentum and continue exploring new molecules! If you encounter any challenges on molecular ratings or have any questions, please do not hesitate to reach out.
